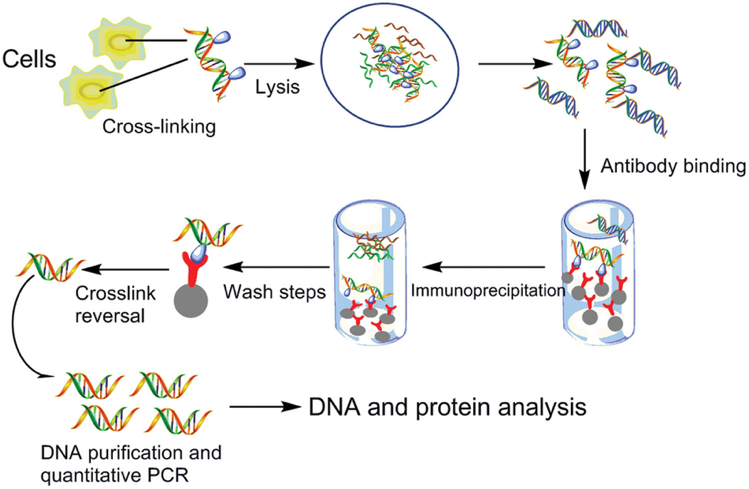
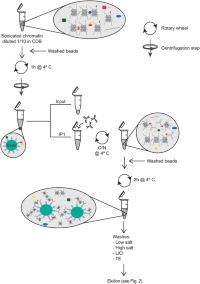
ChIP and re-ChIP assays. A, top panel, map of the ERV-9 LTR. E, the LTR... | Download Scientific Diagram
Enhancer-mediated enrichment of interacting JMJD3–DDX21 to ENPP2 locus prevents R-loop formation and promotes transcription

Reverse Chromatin Immunoprecipitation (R-ChIP) enables investigation of the upstream regulators of plant genes | Communications Biology

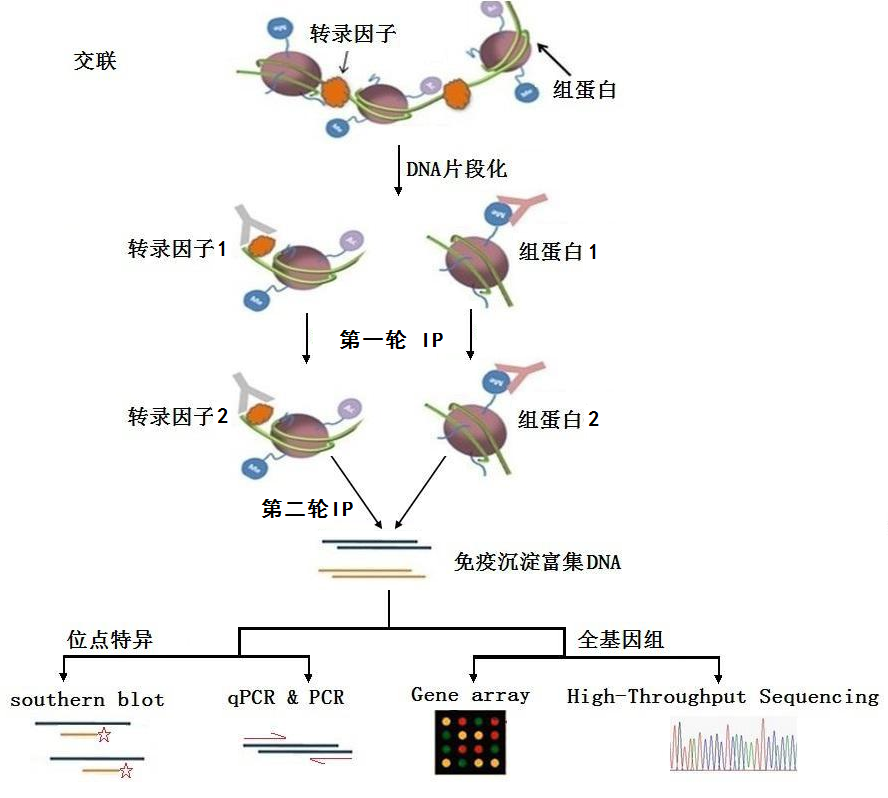
reChIP-seq reveals widespread bivalency of H3K4me3 and H3K27me3 in CD4+ memory T cells | Nature Communications

Figure 6 from SUMOylation of the Farnesoid X Receptor (FXR) Regulates the Expression of FXR Target Genes* | Semantic Scholar

Re-recordable Sound Chip Module, DIY Greeting Card Chip, 120 Seconds Music Sound Voice Recording Player Chip Module for DIY Cards/Toys: Amazon.co.uk: Stationery & Office Supplies

Chip originale di re di JMD per il bambino pratico per 46/4C/4D/G/T5 Chip 10 pz/lotto In azione - AliExpress

NRP 51-RE (TRAY) | netX51 Chip-Carrier con memoria aggiuntiva - CC-Link IE Field Basic Slave | hilscher.com
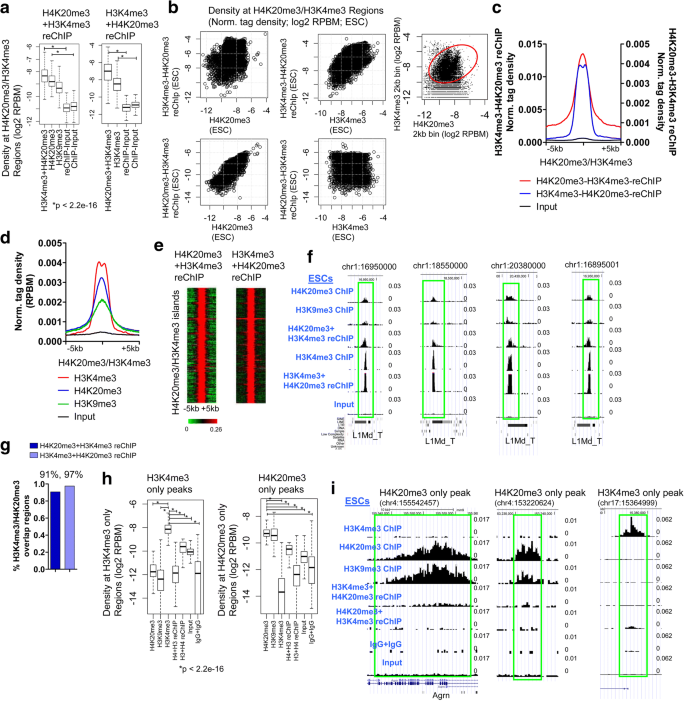
H4K20me3 co-localizes with activating histone modifications at transcriptionally dynamic regions in embryonic stem cells | BMC Genomics | Full Text